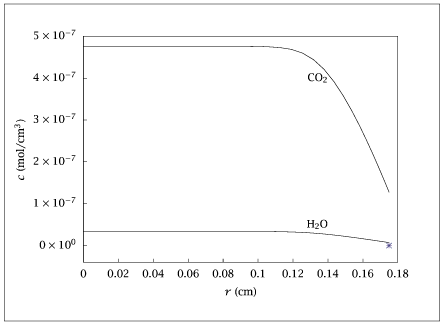
Figure 7.24:
Concentration profiles of products.
Code for Figure 7.24
Text of the GNU GPL.
main.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164 | % Copyright (C) 2001, James B. Rawlings and John G. Ekerdt
%
% This program is free software; you can redistribute it and/or
% modify it under the terms of the GNU General Public License as
% published by the Free Software Foundation; either version 2, or (at
% your option) any later version.
%
% This program is distributed in the hope that it will be useful, but
% WITHOUT ANY WARRANTY; without even the implied warranty of
% MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU
% General Public License for more details.
%
% You should have received a copy of the GNU General Public License
% along with this program; see the file COPYING. If not, write to
% the Free Software Foundation, 59 Temple Place - Suite 330, Boston,
% MA 02111-1307, USA.
%
% Compute the pellet concentration profiles.
%
% Method can address:
%
% multi-component, multi-reaction, arbitrary kinetics
% reaction-diffusion problem in pellet, accurate profiles
% flux and/or value boundary conditions at pellet exterior
% energy balance in pellet and fluid
% pressure drop and equation of state for fluid
% sphere, semi-infinite cylinder or slab geometries for pellet
% axial dispersion in the bed
% transient problem
%
% Method does not address:
%
% radial profiles in bed
% non-uniform pellet exterior
%
% 5/07/02
%
% added log transformation; z=log(c)
% 12/ 13/03
%
% jbr
%
% Revised 8/14/2018
%
% units: mol, cm, sec, K
%
% Simulate pellet with parameters from Co, Cavendish, Hegedus.
%
% C0 + 1/2 O_2 --> CO_2
%
% C_3H_6 + 9/2 O_2 --> 3 CO_2 + 3 H_2O
%
% A=CO B=O_2 C=C_3H_6 D=CO_2 E=H_2O
%
% r_1 = k_1 c_A c_B / (1+K_A c_A + K_C c_C)^2
% r_2 = k_2 c_C c_B / (1+K_A c_A + K_C c_C)^2
p = struct();
p.Rg = 8.314; % J/K mol
p.P = 1.013e5; % N/m^2
p.T = 550; % K
p.cf.tot = p.P/(p.Rg*p.T)*1e-6; % mol/cm^3
p.Rp = 0.175; % cm radius of catalyst particle
p.cf.a = p.cf.tot*0.02;
p.cf.b = p.cf.tot*0.03;
p.cf.c = p.cf.tot*5e-4;
p.k.k10 = 6.802e16*2.6e6*80/100*0.05/100; %mol/cm^3 s
p.k.k20 = 1.416e18*2.6e6*80/100*0.05/100; %mol/cm^3 s
p.k.E1 = 13108; %K
p.k.E2 = 15109; %K
p.k.Ka0 = 8.099e6; % cm^3/mol
p.k.Kc0 = 2.579e8; % cm^3/mol
p.k.Ea = -409; %K
p.k.Ec = 191; %K
p.D.a = 0.0487; % cm^2/s
p.D.b = 0.0469; % cm^2/s
p.D.c = 0.0487; % cm^2/s
p.km.a = 0.4*9.76; % cm/s
p.km.b = 0.4*10.18; % cm/s
p.km.c = 0.4*9.76; % cm/s
% collocation
p.npts = 40;
[p.col.R p.col.A p.col.B p.col.Q] = colloc(p.npts-2, 'left', 'right');
p.col.R = p.col.R*p.Rp;
p.col.A = p.col.A/p.Rp;
p.col.B = p.col.B/(p.Rp*p.Rp);
p.col.Q = p.col.Q*p.Rp;
p.col.Aint = p.col.A(2:p.npts-1,:);
p.col.Bint = p.col.B(2:p.npts-1,:);
p.col.Rint = p.col.R(2:p.npts-1);
%
%
%
% find the pellet profile at the fluid conditions
%
zain = log(1e-9*p.cf.a);
zaout = log(0.75*p.cf.a);
za0 = zain + p.col.R/p.Rp*(zaout - zain);
zbin = log(0.75*p.cf.b);
zbout = log(p.cf.b);
zb0 = zbin + p.col.R/p.Rp*(zbout - zbin);
zcin = log(1e-6*p.cf.c);
zcout = log(p.cf.c);
zc0 = zcin + p.col.R/p.Rp*(zcout - zcin);
z0 = [za0; zb0; zc0];
opts = optimset ('MaxFunEvals', 20*numel(z0), 'MaxIter', 20*numel(z0), ...
'TolX', sqrt(eps), 'TolFun', sqrt(eps));
[z,fval,info] = fsolve(@(x) pellet(x, p), z0, opts);
[dcadr dcbdr dccdr] = dcdr_calc(z, p);
za = z(1:p.npts);
zb = z(p.npts+1:2*p.npts);
zc = z(2*p.npts+1:3*p.npts);
ca = exp(za);
cb = exp(zb);
cc = exp(zc);
% compute other products concentrations
p.km.d = p.km.a;
p.km.e = p.km.b;
p.D.d = p.D.a;
p.D.e = p.D.b;
p.cf.d = 0;
p.cf.e = 0;
dcddr = (-p.D.a*dcadr(p.npts) - 3*p.D.c*dccdr(p.npts))/p.D.d;
dcedr = (-3*p.D.c*dccdr(p.npts))/p.D.e;
cdR = p.cf.d - p.D.d*dcddr/p.km.d;
ceR = p.cf.e - p.D.e*dcedr/p.km.e;
concd = (p.D.a*(ca(p.npts) - ca) + 3*p.D.c*(cc(p.npts) - cc))/p.D.d + cdR;
ce = (3*p.D.c*(cc(p.npts)-cc))/p.D.e + ceR;
table = [p.col.R ca cb cc concd ce dcadr dcbdr dccdr];
table_2 = [p.Rp p.cf.a p.cf.b p.cf.c p.cf.d p.cf.e];
save multi_log.dat table table_2;
if (~ strcmp (getenv ('OMIT_PLOTS'), 'true')) % PLOTTING
subplot (3, 1, 1);
plot (table(:,1), table(:,2:4), table_2(:,1), table_2(:,2:4));
% TITLE
subplot (3, 1, 2);
semilogy (table(:,1), table(:,2:4), table_2(:,1), table_2(:,2:4));
% TITLE
subplot (3, 1, 3);
plot (table(:,1), table(:,5:6), table_2(:,1), table_2(:,5:6));
% TITLE
end % PLOTTING
|
pellet.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52 | function retval = pellet(x, p)
za = x(1:p.npts);
zb = x(p.npts+1:2*p.npts);
zc = x(2*p.npts+1:3*p.npts);
ca = exp(za);
cb = exp(zb);
cc = exp(zc);
k1 = p.k.k10*exp(-p.k.E1/p.T);
k2 = p.k.k20*exp(-p.k.E2/p.T);
Ka = p.k.Ka0*exp(-p.k.Ea/p.T);
Kc = p.k.Kc0*exp(-p.k.Ec/p.T);
den = (1 + Ka*ca + Kc*cc).^2;
r1 = k1.*ca.*cb./den;
r2 = k2*cc.*cb./den;
Ra = - r1;
Rb = - 1/2*r1 - 9/2*r2;
Rc = - r2;
% component A
ip = 1;
retval(ip) = p.col.A(1,:)*za;
caint = ca(2:p.npts-1);
retval(ip+1:ip+p.npts-2) = p.col.Bint*za + p.col.Aint*za.*...
(p.col.Aint*za + 2./p.col.Rint) + ...
Ra(2:p.npts-1)./(p.D.a*caint);
dzadr = p.col.A(p.npts,:)*za;
retval(ip+p.npts-1) = p.D.a*dzadr - p.km.a*(p.cf.a/ca(p.npts) - 1);
% component B
ip = p.npts + 1;
cbint = cb(2:p.npts-1);
retval(ip) = p.col.A(1,:)*zb;
retval(ip+1:ip+p.npts-2) = p.col.Bint*zb + p.col.Aint*zb.*...
(p.col.Aint*zb + 2./p.col.Rint) + ...
Rb(2:p.npts-1)./(p.D.b*cbint);
dzbdr = p.col.A(p.npts,:)*zb;
retval(ip+p.npts-1) = p.D.b*dzbdr - p.km.b*(p.cf.b/cb(p.npts) - 1);
% component C
ip = 2*p.npts + 1;
ccint = cc(2:p.npts-1);
retval(ip) = p.col.A(1,:)*zc;
retval(ip+1:ip+p.npts-2) = p.col.Bint*zc + p.col.Aint*zc.*...
(p.col.Aint*zc + 2./p.col.Rint) + ...
Rc(2:p.npts-1)./(p.D.c*ccint);
dzcdr = p.col.A(p.npts,:)*zc;
retval(ip+p.npts-1) = p.D.c*dzcdr - p.km.c*(p.cf.c/cc(p.npts) - 1);
dcadr = dzadr.*ca;
dcbdr = dzbdr.*cb;
dccdr = dzcdr.*cc;
|
dcdr_calc.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16 | function [dcadr, dcbdr, dccdr] = dcdr_calc(x, p)
za = x(1:p.npts);
zb = x(p.npts+1:2*p.npts);
zc = x(2*p.npts+1:3*p.npts);
ca = exp(za);
cb = exp(zb);
cc = exp(zc);
dzadr = p.col.A(p.npts,:)*za;
dzbdr = p.col.A(p.npts,:)*zb;
dzcdr = p.col.A(p.npts,:)*zc;
dcadr = dzadr.*ca;
dcbdr = dzbdr.*cb;
dccdr = dzcdr.*cc;
|
