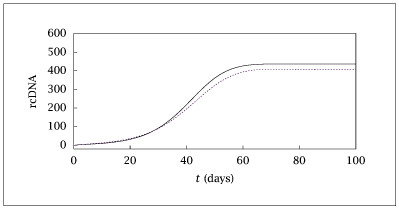
Figure 9.27:
Species rcDNA versus time for hepatitis B virus model.
Code for Figure 9.27
Text of the GNU GPL.
main.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163 | % jbr, elh, 11/28/01
%
% A = cccDNA, B = rcDNA, C = env
%
% Reactions
% 1: A -> A + B
% 2: B -> A
% 3: A -> A + C
% 4: A -> 0 (degradation)
% 5: C -> 0 (degradation)
% 6: B + C -> 0 (secreted virus)
% converted to use parest, jbr, 4/24/2007
% converted to use paresto (casadi), jbr, 4/13/2018
% updated to paresto structs for parameters, jbr, 5/24/2019
% updated to revised paresto, jbr, 6/7/2020
% pass constants with anonymous functions, not as parameters, jbr, 4/3/2021
model=struct();
model.print_level = 0;
%model.nlp_solver_options.ipopt.linear_solver = 'ma27';
model.nlp_solver_options.ipopt.mumps_scaling = 0;
model.transcription = 'shooting';
model.x = {'ca', 'cb', 'cc'};
model.p = {'k1', 'k2', 'k3', 'k4', 'k5', 'k6'};
model.d = {'ma', 'mb', 'mc'};
model.ode = @hbv_rxs;
% measurement weights
mweight = sqrt([1; 1e-2; 1e-4]);
w.a = mweight(1); w.b = mweight(2); w.c = mweight(3);
model.lsq = @(t, y, p) lsq_weighted(t, y, p, w);
% Set the reaction rate constant vector kr; use log transformation
krac = [2; 0.025; 1000; 0.25; 1.9985; 7.5E-6];
thetaac = log10(krac);
for i = 1:numel(model.p)
fn = model.p{i};
p.(fn) = thetaac(i);
end
% output times
tfinal = 100;
ndata = 51;
tdata = linspace(0, tfinal, ndata)';
model.tout = tdata;
pe = paresto(model);
% Set the initial condition
small = 0;
x0_ac = [1; small; small];
y_ac = pe.simulate(zeros(3, 1), x0_ac, p);
randn('seed',1);
R = diag([0.1 0.1 0.1])^2;
noise = sqrt(R)*randn(size(y_ac));
% proportional error
y_noisy = y_ac .* (1 + noise);
y_noisy = max(y_noisy, 0);
% initial guess for rate constants, page 544, edition 2.2.
logkrinit = [0.80, -1.13, 3.15, -0.77, -0.16, -5.46]';
p_init = [logkrinit];
% outputs for the initial guess
y_init = pe.simulate(zeros(3, 1), x0_ac, p_init);
% initialize all parameters
% index of estimated rate constants
del = 1;
% Bounds for estimated parameters
for i = 1:numel(model.p)
fn = model.p{i};
theta0.(fn) = logkrinit(i);
lb.(fn) = theta0.(fn) - del;
ub.(fn) = theta0.(fn) + del;
end
for i = 1:numel(model.x)
fn = model.x{i};
theta0.(fn) = x0_ac(i);
lb.(fn) = theta0.(fn);
ub.(fn) = theta0.(fn);
end
[est, y, p] = pe.optimize(y_noisy, theta0, lb, ub);
conf = pe.confidence(est, 0.95);
disp('Initial guess')
disp(theta0)
disp('Optimal parameters')
disp(est.theta)
disp('Confidence intervals')
disp(conf.bbox)
disp('True parameters')
disp(thetaac)
% check eigenvalues/eigenvectors for badly determined parameter
% directions
[u, lam] = eig(conf.H);
%
% watch out, eigenvalues may be unordered;
% run sort so i(1) will be the index of the smallest eigenvalue;
% still have a +/- sign ambiguity on u(:,i(1)), but thetap with either
% sign on u(:,i(1)) should give the same fit to the data
[s,i] = sort(diag(lam));
logkrp = cell2mat(struct2cell(est.theta)) + 0.5*u(:,i(1));
% simulate with parameters perturbed in weak direction
pp = [logkrp];
yp = pe.simulate(zeros(3,1), x0_ac, pp);
store1 = [model.tout, [y.ca;y.cb;y.cc]', y_init', yp'];
store2 = [model.tout, y_noisy'];
save hbv_det.dat store2 store1;
if (~ strcmp (getenv ('OMIT_PLOTS'), 'true')) % PLOTTING
subplot (3, 2, 1);
plot (model.tout, y_noisy(1,:), '+', model.tout, [y.ca; y_init(1,:)]);
axis ([0, 100, 0, 60]);
% TITLE hbv_det_cccdna
subplot (3, 2, 3);
plot (model.tout, y_noisy(2,:), '+', model.tout, [y.cb; y_init(2,:)]);
axis ([0, 100, 0, 600]);
% TITLE hbv_det_rcdna
subplot (3, 2, 5);
plot (model.tout, y_noisy(3,:), '+', model.tout, [y.cc; y_init(3,:)]);
axis ([0, 100, 0, 30000]);
% TITLE hbv_det_env
subplot (3, 2, 2);
plot (model.tout, y_noisy(1,:), '+', model.tout, [y.ca; yp(1,:)]);
axis ([0, 100, 0, 60]);
% TITLE hbv_det_cccdnap
subplot (3, 2, 4);
plot (model.tout, y_noisy(2,:), '+', model.tout, [y.cb; yp(2,:)]);
axis ([0, 100, 0, 600]);
% TITLE hbv_det_rcndap
subplot (3, 2, 6);
plot (model.tout, y_noisy(3,:), '+', model.tout, [y.cc; yp(3,:)]);
axis ([0, 100, 0, 30000]);
% TITLE hbv_det_envp
end % PLOTTING
|
hbv_rxs.m
| function rhs = hbv_rxs(t, y, p)
kr = [ 10.^p.k1, ...
10.^p.k2, ...
10.^p.k3, ...
10.^p.k4, ...
10.^p.k5, ...
10.^p.k6 ];
rhs = {kr(2)*y.cb - kr(4)*y.ca, ...
kr(1)*y.ca - kr(2)*y.cb - kr(6)*y.cb*y.cc, ...
kr(3)*y.ca - kr(5)*y.cc - kr(6)*y.cb*y.cc};
|
lsq_weighted.m
| function retval = lsq_weighted(t, y, p, w)
retval = {w.a*(y.ca-y.ma), w.b*(y.cb-y.mb), w.c*(y.cc-y.mc)};
|
