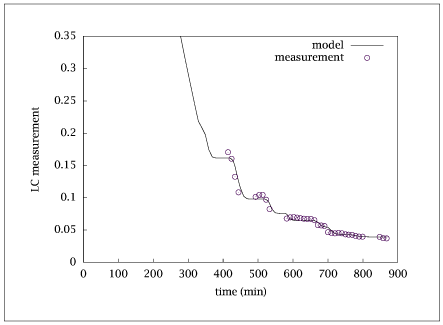
Figure 9.32:
Comparison of data to model with optimal parameters.
Code for Figure 9.32
Text of the GNU GPL.
main.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195 | %
% We have the ODE
% dot(VR) = Qf
% dot(nA) = -k1*nA*nB/VR
% dot(nB) = Qf*cBf - nB*(k1*nA + k2*nC)/VR
% dot(nC) = nB*(k1*nA - k2*nC)/VR
% dot(nD) = k2*nC*nB/VR
%
%
% with:
% States: x = [VR, nA, nB, nC, nD]
% Qf: Volumetric flowrate of base
% cBf: Feed concentration of B
%
% Initial conditions: x(0) = [VR0, nA0, 0, 0, 0]
% Unknown parameters: k1, k2
% Output function: y = nC/(cC + 2*nD)
%
% converted bvsm.m to work with paresto.m
%
% Joel Andersson and jbr, 4/22/2018
%
% Model
% This m-file loads data file 'lc.dat'.
% This m-file loads data file 'flow.dat'.
model = struct;
model.transcription = 'shooting';
model.x = {'VR', 'nA', 'nB', 'nC', 'nD'};
model.p = {'k1', 'k2', 'cBf'};
model.nlp_solver_options.ipopt.mumps_scaling = 0;
% Dependent variables with definitions
model.y = {'lc'};
model.h = @(t, v, p) { 1 / (1 + 2*v.nD/max(v.nC, 1e-6))}; % avoid divide-by-zero
% Data and measurements
model.d = {'Qf', 'lc_m'};
% ODE right-hand-side
model.ode = @(t, v, p) {v.Qf,...
-p.k1*v.nA*v.nB/v.VR,...
v.Qf*p.cBf - v.nB*(p.k1*v.nA + p.k2*v.nC)/v.VR,...
v.nB*(p.k1*v.nA - p.k2*v.nC)/v.VR,...
p.k2*v.nC*v.nB/v.VR};
% Relative least squares objective function
model.lsq = @(t, y, p) {y.lc_m/y.lc - 1};
% Load data
teaf = 0.00721;
teaden = 0.728;
flow = load('flow.dat');
lc = load('lc.dat');
tQf = [0. ; flow(:,1)];
Qf = [0. ; flow(:,2)./teaden];
tlc = lc(:,1);
lc = lc(:,2);
% Get all time points occuring in either tlc or tflow
% Grid used in Book
% ntimes = 200;
% tlin = linspace (0, tQf(end), ntimes)';
% [tout,~,ic] = unique([tQf; tlc; tlin]);
% Qf_ind = ic(1:numel(tQf));
% lc_ind = ic(numel(tQf)+1:numel(tQf)+numel(tlc));
% faster grid; lose a little resolution in n_B(t) plot
[tout,~,ic] = unique([tQf; tlc]);
Qf_ind = ic(1:numel(tQf));
lc_ind = ic(numel(tQf)+1:end);
% Interpolate lcmeas and Qf to this new grid
Qf = interp1(tQf, Qf, tout, 'previous');
lc_m = interp1(tlc, lc, tout, 'previous');
N = size(Qf,1);
% Replace NaNs with zeros
Qf(isnan(Qf)) = 0.;
lc_m(isnan(lc_m)) = 0.;
% Initial volume
VR0 = 2370;
% Options
model.tout = tout';
model.lsq_ind = lc_ind'; % only include lc_ind in objective
% Create a paresto instance
pe = paresto(model);
% Solution in the book
nA0 = 2.35;
k1 = 2500;
k2 = 1250;
% Options
model.tout = tout';
model.lsq_ind = lc_ind'; % only include lc_ind in objective
% Create a paresto instance
pe = paresto(model);
% Initial guess for parameters
theta0 = struct;
theta0.k1 = k1;
theta0.k2 = 0.9*k2;
theta0.cBf = teaf;
% Initial guess for initial conditions
theta0.VR = VR0;
theta0.nA = 1.1*nA0;
theta0.nB = 0;
theta0.nC = 0;
theta0.nD = 0;
% Lower bounds for parameters
lb = theta0;
lb.k1 = 0.5*theta0.k1;
lb.k2 = 0.5*theta0.k2;
lb.cBf = theta0.cBf;
lb.VR = theta0.VR;
lb.nA = 0.5*theta0.nA;
lb.nB = theta0.nB;
lb.nC = theta0.nC;
lb.nD = theta0.nD;
% Upper bounds for parameters
ub = theta0;
ub.k1 = 1.5*theta0.k1;
ub.k2 = 1.5*theta0.k2;
ub.cBf = theta0.cBf;
ub.VR = theta0.VR;
ub.nA = 1.5*theta0.nA;
ub.nB = theta0.nB;
ub.nC = theta0.nC;
ub.nD = theta0.nD;
% Estimate parameters
more off
[est,v,p] = pe.optimize([Qf'; lc_m'], theta0, lb, ub);
%est_ind = [1,2,3]; % index of estimated parameters
% Also calculate confidence intervals with 95 % confidence
conf = pe.confidence(est, 0.95);
disp('Estimated parameters')
disp(est.theta)
disp('Bounding box intervals')
disp(conf.bbox)
data = struct();
data.model = [model.tout', est.x', v.lc'];
data.measurement = [tlc, lc];
gnuplotsave('bvsm.dat', data);
if (~ strcmp (getenv ('OMIT_PLOTS'), 'true')) % PLOTTING
clf
subplot(2,2,1)
hold on
plot(model.tout, v.nA)
plot(model.tout, v.nC)
plot(model.tout, v.nD)
legend({'n_A', 'n_C', 'n_D'});
xlabel('time (min)')
ylabel('Amount of substance (kmol)')
title('Amount of substance of species A, C and D versus time')
subplot(2,2,2)
hold on
plot(model.tout, v.nB)
legend({'n_B'});
xlabel('time (min)')
ylabel('Amount of substance (kmol)')
title('Amount of substance of species B versus time')
subplot(2,2,3)
stairs(model.tout, v.Qf)
xlabel('time (min)')
ylabel('flowrate (kg/min)')
title('Base addition rnate')
subplot(2,2,4)
hold on
plot(model.tout, v.lc)
plot(tlc, lc, 'o')
ylim([0, 2*max(lc)])
legend({'model', 'measurement'});
xlabel('time (min)')
title('LC measurement')
end % PLOTTING
|
lc.dat
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35 | 414 0.17050
424 0.16040
434 0.13250
444 0.10840
493 0.10140
503 0.10420
513 0.10450
523 0.09700
533 0.08240
582 0.06780
592 0.06970
602 0.07020
612 0.06920
621 0.06840
631 0.06750
641 0.06730
651 0.06730
661 0.06540
671 0.05770
681 0.05710
690 0.05630
700 0.04660
710 0.04550
720 0.04490
730 0.04560
740 0.04500
750 0.04340
759 0.04280
769 0.04260
779 0.04090
789 0.03990
799 0.03970
848 0.03940
858 0.03740
868 0.03710
|
flow.dat
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87 | 9.000000 9.7817838E-02 4.786000
19.00000 1.290601 14.21800
29.00000 1.262779 13.99800
39.00000 1.240016 13.81800
49.00000 1.215736 13.62600
59.00000 1.187156 13.40000
69.00000 1.160599 13.19000
79.00000 0.5545961 8.398001
89.00000 1.109508 12.78600
99.00000 1.361925 14.78200
109.0000 1.332333 14.54800
119.0000 1.294648 14.25000
129.0000 1.271885 14.07000
139.0000 1.244569 13.85400
149.0000 1.220794 13.66600
159.0000 1.197778 13.48400
169.0000 1.170463 13.26800
179.0000 1.150988 13.11400
189.0000 1.128983 12.94000
199.0000 1.105968 12.75800
209.0000 1.092563 12.65200
219.0000 1.360155 14.76800
229.0000 1.344473 14.64400
239.0000 1.329045 14.52200
249.0000 1.305524 14.33600
259.0000 1.275426 14.09800
269.0000 1.259239 13.97000
279.0000 1.244063 13.85000
289.0000 1.219783 13.65800
299.0000 1.191961 13.43800
309.0000 1.173751 13.29400
319.0000 1.158322 13.17200
329.0000 0.3163429 6.514000
339.0000 0.5495375 8.358001
349.0000 1.224841 13.69800
359.0000 0.3228684 6.565600
369.0000 9.4845297E-04 4.020000
379.0000 4.6794413E-04 4.016201
389.0000 5.4383278E-04 4.016800
399.0000 4.9326418E-04 4.016400
409.0000 3.6680698E-04 4.015400
419.0000 0.2701340 6.148601
429.0000 1.185891 13.39000
439.0000 0.9299334 11.36600
449.0000 0.5259147 8.171202
459.0000 8.9793204E-04 4.019600
469.0000 8.7256433E-04 4.019400
479.0000 7.9672335E-04 4.018800
489.0000 4.9319270E-04 4.016400
499.0000 4.6799183E-04 4.016200
509.0000 3.6678315E-04 4.015400
519.0000 0.2330809 5.855600
529.0000 0.7784327 10.16800
539.0000 7.5244429E-03 4.072000
549.0000 7.5244186E-03 4.072000
559.0000 5.7539940E-03 4.058000
569.0000 8.0302954E-03 4.076001
579.0000 0.5019882 7.982001
589.0000 -3.0983209E-03 3.988000
599.0000 9.4850064E-04 4.020000
609.0000 -6.3252446E-05 4.012000
619.0000 -1.3278484E-03 4.002000
629.0000 -4.6158792E-03 3.976000
639.0000 -6.3204767E-05 4.012000
649.0000 -2.0865917E-03 3.996000
659.0000 0.1752122 5.398000
669.0000 0.3024322 6.404000
679.0000 4.9952744E-03 4.052001
689.0000 3.9835931E-03 4.044001
699.0000 0.5114223 8.056601
709.0000 3.4151078E-04 4.015200
719.0000 3.1619071E-04 4.015000
729.0000 1.1386871E-04 4.013400
739.0000 6.3300133E-05 4.013000
749.0000 3.4151078E-04 4.015200
759.0000 0.1059619 4.850400
769.0000 6.7019463E-04 4.017800
779.0000 6.8099439E-02 4.551000
789.0000 8.7261200E-04 4.019400
799.0000 9.7372534E-04 4.020200
809.0000 8.7256433E-04 4.019400
819.0000 7.9669955E-04 4.018800
829.0000 7.2083471E-04 4.018200
839.0000 5.6912901E-04 4.017000
849.0000 4.6801567E-04 4.016200
859.0000 5.1856041E-04 4.016600
869.0000 4.4267176E-04 4.016000
|
