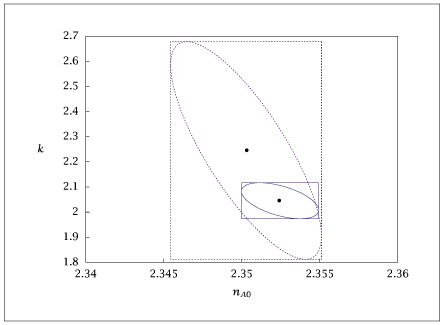
Figure 9.37:
Confidence intervals for reduced model without (dashed) and with (solid) redesigned experiment.
Code for Figure 9.37
Text of the GNU GPL.
main.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296 | %
% We have the reduced ODE model
% dot(VR) = Qf
% dot(eps2) = Qf*cBf/(1 + k (nA0-nBadded+eps2)/(nBadded-2*eps2))
%
%
% with:
% States: x = [VR, eps2] volume and extent of 2nd reaction
% Qf: Volumetric flowrate of base
% cBf: Feed concentration of B
%
% Initial conditions: x(0) = [VR0, 0]
% Unknown parameters: k (= k1/k2), n_A0
% Output function: y = nC/(nC + 2*nD) = 1/(1+2*nD/nC)
%
% jbr, Joel Andersson, 4/18/2018
%
% Model
% This m-file loads data file 'lc.dat'.
% This m-file loads data file 'flow.dat'.
% This m-file loads data file 'lcsim.dat'.
model = struct;
model.transcription = 'shooting';
%model.nlp_solver_options.ipopt.linear_solver = 'ma27';
model.nlp_solver_options.ipopt.mumps_scaling = 0;
% set eps to zero for algebraic model
model.nlp_solver_options.sens_linsol_options.eps = 0;
model.x = {'VR', 'eps2'};
model.p = {'nA0', 'k', 'cBf', 'VR0'};
% Dependent variables with definitions
model.y = {'lc'};
model.h = @lcmeas;
% Data and measurements
model.d = {'Qf', 'lc_m'};
% ODE right-hand-side
model.ode = @reduced_model;
% Relative least squares objective function
model.lsq = @(t, y, p) {y.lc_m/y.lc - 1};
% Load data
teaf = 0.00721;
teaden = 0.728;
flow = load('flow.dat');
lc = load('lc.dat');
tQf = [0. ; flow(:,1)];
Qf = [0. ; flow(:,2)./teaden];
tlc = lc(:,1);
lc = lc(:,2);
% Get all time points occuring in either tlc or tflow
% Grid used in Book
% ntimes = 200;
% tlin = linspace (0, tQf(end), ntimes)';
% [tout,~,ic] = unique([tQf; tlc; tlin]);
% Qf_ind = ic(1:numel(tQf));
% lc_ind = ic(numel(tQf)+1:numel(tQf)+numel(tlc));
% faster grid; lose a little resolution in n_B(t) plot
[tout,~,ic] = unique([tQf; tlc]);
Qf_ind = ic(1:numel(tQf));
lc_ind = ic(numel(tQf)+1:end);
% Interpolate lcmeas and Qf to this new grid
Qf = interp1(tQf, Qf, tout, 'previous');
lc_m = interp1(tlc, lc, tout, 'previous');
N = size(Qf,1);
% Replace NaNs with zeros
Qf(isnan(Qf)) = 0.;
lc_m(isnan(lc_m)) = 0.;
% Initial volume
VR0 = 2370;
% "optimal" values
vrdiclo = 2.3497;
k1k2ratio = 2;
% Options
model.tout = tout';
model.lsq_ind = lc_ind'; % only include lc_ind in objective
% Create a paresto instance
pe = paresto(model);
% Initial guess for parameters
theta0 = struct;
theta0.nA0 = vrdiclo;
theta0.k = k1k2ratio;
theta0.cBf = teaf;
theta0.VR0 = VR0;
% Initial guess for initial conditions
theta0.VR = VR0;
theta0.eps2 = 0;
% Lower bounds for parameters
% We aren't estimating cBf and VR0
lb = theta0;
lb.nA0 = 0.5*theta0.nA0;
lb.k = 0.5*theta0.k;
lb.VR0 = theta0.VR0;
lb.cBf = theta0.cBf;
lb.VR = theta0.VR;
lb.eps2 = theta0.eps2;
% Upper bounds for parameters
ub = theta0;
ub.nA0 = 1.5*theta0.nA0;
ub.k = 1.5*theta0.k;
ub.VR0 = theta0.VR0;
ub.cBf = theta0.cBf;
ub.VR = theta0.VR;
ub.eps2 = theta0.eps2;
% Estimate parameters
[est, v, p] = pe.optimize([Qf'; lc_m'], theta0, lb, ub);
% Also calculate confidence intervals with 95 % confidence
conf = pe.confidence(est, 0.95);
disp('Estimated parameters')
disp(est.theta)
disp('Bounding box intervals')
disp(conf.bbox)
np = numel(est.conf_ind);
ndata = length(tlc);
alpha = 0.95;
Fstat = np*finv(alpha,np,ndata-np);
a = 2*est.f/(ndata-np)*Fstat;
[xx, yy, major, minor, bbox] = ellipse (conf.H, a, 100, [est.theta.nA0; est.theta.k]);
tmp = [xx, yy];
table1 = [model.tout', v.lc'];
table2 = [tlc, lc];
% Estimate parameters again with the early time LC data
%
% Simulated the lc measurement from the beginning of the experiment.
% These data were saved by hand in lcsim.dat with
% k1k2ratio=2, vrdiclo=2.3497;
% 2nd column is without noise, 3rd column is with noise.
%
% prepend simulated data
%
flow = load('flow.dat');
lc = load('lc.dat');
lcsim = load('lcsim.dat');
tQf = [0. ; flow(:,1)];
Qf = [0. ; flow(:,2)./teaden];
tlc = [lcsim(:,1); lc(:,1)];
lc = [lcsim(:,3); lc(:,2)];
% Get all time points occuring in either tlc or tflow
% Grid used in Book
% ntimes = 200;
% tlin = linspace (0, tQf(end), ntimes)';
% [tout,~,ic] = unique([tQf; tlc; tlin]);
% Qf_ind = ic(1:numel(tQf));
% lc_ind = ic(numel(tQf)+1:numel(tQf)+numel(tlc));
% faster grid; lose a little resolution in n_B(t) plot
[tout,~,ic] = unique([tQf; tlc]);
Qf_ind = ic(1:numel(tQf));
lc_ind = ic(numel(tQf)+1:end);
% Interpolate lcmeas and Qf to this new grid
Qf = interp1(tQf, Qf, tout, 'previous');
lc_m = interp1(tlc, lc, tout, 'previous');
N = size(Qf,1);
% Replace NaNs with zeros
Qf(isnan(Qf)) = 0.;
lc_m(isnan(lc_m)) = 0.;
% Initial volume
VR0 = 2370;
% optimal values
vrdiclo = 2.3497;
k1k2ratio = 2;
% Options
model.tout = tout';
model.lsq_ind = lc_ind'; % only include lc_ind in objective
% Create a paresto instance
pesim = paresto(model);
% Lower bounds for parameters
lb = theta0;
lb.nA0 = 0.5*theta0.nA0;
lb.k = 0.5*theta0.k;
% We aren't estimating cBf and VR0
lb.VR = [theta0.VR, -inf(1,N-1)]; %zeros(1,N-1)];
lb.eps2 = [theta0.eps2, -inf(1,N-1)]; %zeros(1,N-1)];
% Upper bounds for parameters
ub = theta0;
ub.nA0 = 1.5*theta0.nA0;
ub.k = 1.5*theta0.k;
ub.VR = [theta0.VR, inf(1,N-1)];
ub.eps2 = [theta0.eps2, inf(1,N-1)];
% Estimate parameters
[estsim, v, p] = pesim.optimize([Qf'; lc_m'], theta0, lb, ub);
% Also calculate confidence intervals with 95 % confidence
confsim = pesim.confidence(estsim, 0.95);
disp('Estimated parameters')
disp(estsim.theta)
disp('Bounding box intervals')
disp(confsim.bbox)
% compute the rest of the states from the reduced model
Badded = (v.VR - p.VR0)*p.cBf;
nD = v.eps2;
nC = Badded - 2*nD;
nB = zeros(size(Badded));
eps1 = Badded-v.eps2;
nA = p.nA0 - Badded + v.eps2;
deps2dt = v.Qf*p.cBf / (1. + p.k*(p.nA0 - Badded + v.eps2)/(Badded - 2*v.eps2));
ndata = length(tlc);
Fstat = np*finv(alpha, np, ndata-np);
a = 2*estsim.f/(ndata-np)*Fstat;
[xxex, yyex, major, minor, bboxex] = ...
ellipse (confsim.H, a, 100, [estsim.theta.nA0; estsim.theta.k]);
tmpex = [xxex, yyex];
table1ex = [model.tout', v.lc'];
table2ex = [tlc, lc];
if (~ strcmp (getenv ('OMIT_PLOTS'), 'true')) % PLOTTING
subplot(2,2,1)
hold on
plot(model.tout, nA)
plot(model.tout, nC)
plot(model.tout, nD)
legend({'n_A', 'n_C', 'n_D'});
xlabel('time (min)')
ylabel('Amount of substance (kmol)')
title('Amount of substance of species A, C and D versus time')
subplot(2,2,2)
hold on
plot(model.tout, nB)
legend({'n_B'});
xlabel('time (min)')
ylabel('Amount of substance (kmol)')
title('Amount of substance of species B versus time')
subplot(2,2,3)
stairs(model.tout, v.Qf)
xlabel('time (min)')
ylabel('flowrate (kg/min)')
title('Base addition rate')
subplot(2,2,4)
hold on
plot(model.tout, v.lc, tlc, lc, 'o')
%ylim([0, 2*max(lc)])
legend({'model', 'measurement'});
xlabel('time (min)')
title('LC measurement')
figure()
plot(xx, yy, bbox(:,1), bbox(:,2), est.theta.nA0, est.theta.k, 'x', ...
xxex, yyex, bboxex(:,1), bboxex(:,2), estsim.theta.nA0, estsim.theta.k, 'o')
end % PLOTTING
save bvsm_red.dat table1 table2 bbox tmp table1ex table2ex bboxex tmpex;
|
lc.dat
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35 | 414 0.17050
424 0.16040
434 0.13250
444 0.10840
493 0.10140
503 0.10420
513 0.10450
523 0.09700
533 0.08240
582 0.06780
592 0.06970
602 0.07020
612 0.06920
621 0.06840
631 0.06750
641 0.06730
651 0.06730
661 0.06540
671 0.05770
681 0.05710
690 0.05630
700 0.04660
710 0.04550
720 0.04490
730 0.04560
740 0.04500
750 0.04340
759 0.04280
769 0.04260
779 0.04090
789 0.03990
799 0.03970
848 0.03940
858 0.03740
868 0.03710
|
flow.dat
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87 | 9.000000 9.7817838E-02 4.786000
19.00000 1.290601 14.21800
29.00000 1.262779 13.99800
39.00000 1.240016 13.81800
49.00000 1.215736 13.62600
59.00000 1.187156 13.40000
69.00000 1.160599 13.19000
79.00000 0.5545961 8.398001
89.00000 1.109508 12.78600
99.00000 1.361925 14.78200
109.0000 1.332333 14.54800
119.0000 1.294648 14.25000
129.0000 1.271885 14.07000
139.0000 1.244569 13.85400
149.0000 1.220794 13.66600
159.0000 1.197778 13.48400
169.0000 1.170463 13.26800
179.0000 1.150988 13.11400
189.0000 1.128983 12.94000
199.0000 1.105968 12.75800
209.0000 1.092563 12.65200
219.0000 1.360155 14.76800
229.0000 1.344473 14.64400
239.0000 1.329045 14.52200
249.0000 1.305524 14.33600
259.0000 1.275426 14.09800
269.0000 1.259239 13.97000
279.0000 1.244063 13.85000
289.0000 1.219783 13.65800
299.0000 1.191961 13.43800
309.0000 1.173751 13.29400
319.0000 1.158322 13.17200
329.0000 0.3163429 6.514000
339.0000 0.5495375 8.358001
349.0000 1.224841 13.69800
359.0000 0.3228684 6.565600
369.0000 9.4845297E-04 4.020000
379.0000 4.6794413E-04 4.016201
389.0000 5.4383278E-04 4.016800
399.0000 4.9326418E-04 4.016400
409.0000 3.6680698E-04 4.015400
419.0000 0.2701340 6.148601
429.0000 1.185891 13.39000
439.0000 0.9299334 11.36600
449.0000 0.5259147 8.171202
459.0000 8.9793204E-04 4.019600
469.0000 8.7256433E-04 4.019400
479.0000 7.9672335E-04 4.018800
489.0000 4.9319270E-04 4.016400
499.0000 4.6799183E-04 4.016200
509.0000 3.6678315E-04 4.015400
519.0000 0.2330809 5.855600
529.0000 0.7784327 10.16800
539.0000 7.5244429E-03 4.072000
549.0000 7.5244186E-03 4.072000
559.0000 5.7539940E-03 4.058000
569.0000 8.0302954E-03 4.076001
579.0000 0.5019882 7.982001
589.0000 -3.0983209E-03 3.988000
599.0000 9.4850064E-04 4.020000
609.0000 -6.3252446E-05 4.012000
619.0000 -1.3278484E-03 4.002000
629.0000 -4.6158792E-03 3.976000
639.0000 -6.3204767E-05 4.012000
649.0000 -2.0865917E-03 3.996000
659.0000 0.1752122 5.398000
669.0000 0.3024322 6.404000
679.0000 4.9952744E-03 4.052001
689.0000 3.9835931E-03 4.044001
699.0000 0.5114223 8.056601
709.0000 3.4151078E-04 4.015200
719.0000 3.1619071E-04 4.015000
729.0000 1.1386871E-04 4.013400
739.0000 6.3300133E-05 4.013000
749.0000 3.4151078E-04 4.015200
759.0000 0.1059619 4.850400
769.0000 6.7019463E-04 4.017800
779.0000 6.8099439E-02 4.551000
789.0000 8.7261200E-04 4.019400
799.0000 9.7372534E-04 4.020200
809.0000 8.7256433E-04 4.019400
819.0000 7.9669955E-04 4.018800
829.0000 7.2083471E-04 4.018200
839.0000 5.6912901E-04 4.017000
849.0000 4.6801567E-04 4.016200
859.0000 5.1856041E-04 4.016600
869.0000 4.4267176E-04 4.016000
|
lcsim.dat
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73 | 9 0.999999811053562 1.01722259342699
11 0.999587658944779 0.99385414630478
19 0.997925636447302 1.0063033273586
22 0.989776227519767 0.973524345205085
29 0.97073863708603 0.951277459498521
33 0.96009379707585 0.955916127193075
39 0.944126388142496 0.933176658461481
44 0.931059995373657 0.912129208525589
49 0.917993531731551 0.924765345958655
55 0.902620794363343 0.929423720426881
59 0.892372164807407 0.895315152189819
66 0.874858939641787 0.878874365681482
69 0.867353293988678 0.887610914800141
77 0.847785922726378 0.848974596028909
79 0.842894103481712 0.848985755994263
88 0.83237481405589 0.829011613033741
89 0.831205888518126 0.828704133459361
99 0.807823351175219 0.808888562620313
109 0.779121310864896 0.786381406938047
110 0.77631345417554 0.776252149913632
119 0.751042820774906 0.756749889814251
121 0.745585969175354 0.732134962762848
129 0.723758566435087 0.723820015722546
132 0.715717201841988 0.725510391009964
139 0.696954020548539 0.697223175175415
143 0.68646246764278 0.67349142047977
149 0.670725076033402 0.668328166140843
154 0.657861133646678 0.662317372691344
159 0.644997187699813 0.657514340724487
165 0.629851520352139 0.621581418804898
169 0.61975441688066 0.626779273090415
176 0.602487407389775 0.585884236756459
179 0.595087253950168 0.597729143403102
187 0.575681893808323 0.580350965840298
189 0.570830568007055 0.572046209133092
198 0.549416907668722 0.541541840673101
199 0.547037560114431 0.548612747410106
209 0.523729613746048 0.538541249955535
219 0.500704189037897 0.513368588184931
220 0.497837706149487 0.50676216119614
229 0.472039332645793 0.477205527084727
231 0.466372465615925 0.488838142877278
239 0.443705011167859 0.437219925680493
242 0.435302251160462 0.432986828416903
249 0.415695826849098 0.416071875495965
253 0.404690424332785 0.401912894973921
259 0.388182323384162 0.39285382966983
264 0.37474272150017 0.378923137070906
269 0.361303120788802 0.367484106358756
275 0.345380258106417 0.338508666896052
279 0.33476502597149 0.333439320473033
286 0.316412252844235 0.313210164070985
289 0.308546738752642 0.302731294760027
297 0.287981499998565 0.284780383973117
299 0.282840191132878 0.274816315777158
308 0.260231996792122 0.273443081396385
309 0.257719953569755 0.251919518622741
319 0.232983524949703 0.231578003318462
329 0.208572150752592 0.225291260764646
330 0.207905468886441 0.22729540104205
339 0.201905298645643 0.206577487404493
341 0.199589040875074 0.210143334269163
349 0.190323859821764 0.195116540264097
352 0.182579938375238 0.187682610356096
359 0.164510700875708 0.151127109700629
363 0.161788969441419 0.158451310797697
369 0.15770634207812 0.162343283034236
374 0.157696338778412 0.149316304212486
379 0.157686348622417 0.164821037715053
385 0.15768043157753 0.15766516626921
389 0.157676482590873 0.161593237909514
396 0.157668465879349 0.170786320474533
399 0.157665027844229 0.153198631274023
|
lcmeas.m
| function retval = lcmeas(t, v, p)
Badded = (v.VR - p.VR0)*p.cBf;
nD = v.eps2;
nC = Badded - 2*nD;
retval = { 1 / (1 + 2*nD/max(nC, 1e-6))}; % avoid divide-by-zero
|
reduced_model.m
| function xdot = reduced_model(t, v, p)
Badded = max((v.VR - p.VR0)*p.cBf, 1e-6); % avoid divide-by-zero
deps2dt = v.Qf*p.cBf / (1. + p.k*(p.nA0 - Badded + v.eps2)/(Badded - 2*v.eps2));
xdot = {v.Qf, deps2dt};
|
