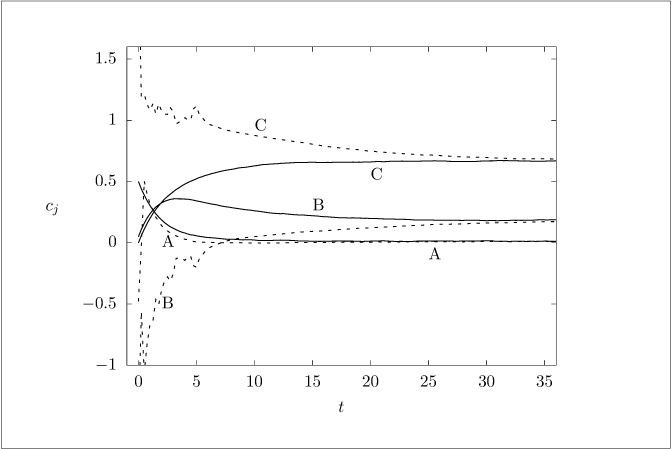
Figure 4.4:
Evolution of the state (solid line) and UKF state estimate (dashed line).
Code for Figure 4.4
Text of the GNU GPL.
main.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14 | % This file simulates the Batch Reactor Example (3.2.1) from Haseltine
% & Rawlings (2005) UKF is implemented to this example and it is shown that it
% fails when a poor prior is used.
data = struct();
% Helper functions.
% Simulate.
fprintf('Simulating without clipping.\n');
data.noclip = doukf(150, false());
fprintf('Simulating with clipping.\n');
data.yesclip = doukf(180, true());
|
batchreactor.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129 | function pars = batchreactor(Nsim)
% pars = batchreactor(Nsim)
%
% Returns a struct of batch reactor parameters to avoid duplication across
% MHE, EKF, and UKF scripts.
narginchk(1, 1);
mpc = import_mpctools();
% Sizes.
Delta = 0.25;
Nx = 3;
Ny = 1;
Nw = Nx;
Nv = Ny;
% Random variable standard deviations.
sig_v = 0.25; % Measurement noise.
sig_w = 0.001; % State noise.
sig_p = 0.5; % Prior.
P = sig_p.^2*eye(Nx);
Q = sig_w.^2*eye(Nw);
R = sig_v.^2*eye(Ny);
% Get model functions.
plant = mpc.getCasadiIntegrator(@batch_model, Delta, [Nx], {'x'});
frk4 = mpc.getCasadiFunc(@(x) batch_model(x), [Nx], {'x'}, {'frk4'}, ...
'rk4', true(), 'Delta', Delta, 'M', 4);
f = mpc.getCasadiFunc(@(x, w) frk4(x) + w, [Nx, Nw], {'x', 'w'}, {'f'});
h = mpc.getCasadiFunc(@measurement, [Nx], {'x'}, {'h'});
% Choose prior.
xhat0 = [1; 0; 4];
x0 = [0.5; 0.05; 0.0];
% Simulate the system to get data.
randn('state', 0);
w = sig_w*randn(Nw, Nsim);
v = sig_v*randn(Nv, Nsim + 1);
xsim = NaN(Nx, Nsim + 1);
xsim(:,1) = x0;
ysim = NaN(Ny, Nsim + 1);
yclean = NaN(Ny, Nsim + 1);
for t = 1:(Nsim + 1)
xsim(:,t) = max(xsim(:,t), 0); % Make sure concentration is nonnegative.
yclean(:,t) = full(h(xsim(:,t)));
ysim(:,t) = yclean(:,t) + v(:,t);
if t <= Nsim
xsim(:,t + 1) = full(plant(xsim(:,t))) + w(:,t);
end
end
t = (0:Nsim)*Delta;
% Get plotting function for convenience.
reactorplot = @reactorplot;
% Package everybody up. Note that this is a quick and dirty approach.
varnames = who();
pars = struct();
for i = 1:length(varnames);
v = varnames{i};
pars.(v) = eval(v);
end
end%function
function dxdt = batch_model(x)
% Nonlinear ODE function.
k1 = 0.5;
km1 = 0.05;
k2 = 0.2;
km2 = 0.01;
cA = x(1);
cB = x(2);
cC = x(3);
rate1 = k1*cA - km1*cB*cC;
rate2 = k2*cB.^2 - km2*cC;
dxdt = [
-rate1;
rate1 - 2*rate2;
rate1 + rate2;
];
end%function
function y = measurement(x)
% Linear measurement function.
RT = 0.0821*400;
y = RT*sum(x);
end%function
function fig = reactorplot(x, xhat, y, yhat, yclean, Delta)
% Make a plot of actual and estimated states.
narginchk(6, 6);
fig = figure();
Nx = 3;
Nt = size(x, 2) - 1;
t = (0:Nt)*Delta;
ax = subplot(2, 1, 2);
hold('on');
plot(t, yclean, '-k', 'DisplayName', 'Actual');
plot(t, yclean, '--c', 'DisplayName', 'Measured');
plot(t, yhat, 'ok', 'DisplayName', 'Estimated');
ylabel('P');
xlabel('t');
legend('Location', 'EastOutside');
set(ax, 'ylim', [10, 35]);
ax = subplot(2, 1, 1);
hold('on');
colors = {'r', 'b', 'g'};
names = {'A', 'B', 'C'};
for i = 1:Nx
plot(t, x(i,:) + 1e-10, ['-', colors{i}], 'DisplayName', ...
sprintf('Actual C_%s', names{i}));
plot(t, xhat(i,:) + 1e-10, ['o', colors{i}], 'DisplayName', ...
sprintf('Estimated C_%s', names{i}));
end
xlabel('t');
ylabel('c_i');
legend('Location', 'EastOutside');
set(ax, 'ylim', [-1, 1.5]);
end%function
|
get_sigma.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21 | function [x_sigma, w_sigma, v_sigma] = get_sigma(Pz, xhat, pars)
% [x_sigma, w_sigma, v_sigma] = get_sigma(Pz, pars)
%
% Returns sigma points obtained from scaled sqrtm(Pz).
narginchk(3, 3);
% Make sure covariance is positive-definite.
[Pzchol, okay] = chol(Pz);
if okay ~= 0
warning('Pz is not positive definite!');
end
% Select sigma points.
sqrtPz = sqrtm((pars.Nz + pars.lambda)*Pz);
zhat = [xhat; zeros(pars.Nw + pars.Nv, 1)];
z_sigma = bsxfun(@plus, [zeros(pars.Nz, 1), sqrtPz, -sqrtPz], zhat);
% Split into pieces.
x_sigma = z_sigma(1:pars.Nx,:);
w_sigma = z_sigma(pars.Nx + (1:pars.Nw),:);
v_sigma = z_sigma(pars.Nx + pars.Nw + (1:pars.Nv),:);
|
ukf_scale.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28 | function [x_sigma, weights] = ukf_scale(x_sigma, pars)
% [x_sigma, weights] = ukf_scale(x_sigma, pars)
%
% Rescales the sigma points and adjusts weights so that each sigma point has
% no negative components.
%
% x_sigma should be a matrix of column vectors with the first vector giving
% the central point (which must always be feasible).
x_center = x_sigma(:,1); % Central point.
x_outer = x_sigma(:,2:end);
% Choose theta to make each point feasible.
dx = abs(bsxfun(@minus, x_outer, x_center));
err = abs(min(x_outer, 0)); % Gets negative part.
err(x_center < 0,:) = 0; % Can't fix indices where x_center is negative.
dx(err == 0) = 1; % Avoid division by zero.
theta = 1 - max(err./dx);
% Rescale outer points.
x_outer = bsxfun(@times, theta, x_outer) ...
+ bsxfun(@times, 1 - theta, x_center);
x_sigma = [x_center, x_outer];
% Calculate weights.
rho = (2*pars.lambda - 1)/(pars.Nsigma - sum(theta));
a = -rho/(2*(pars.Nz + pars.lambda));
b = (1 + rho)/(2*(pars.Nz + pars.lambda));
weights = [0, a*theta]' + b;
|
doukf.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87 | function data = doukf(Nsim, clipping)
% data = doukf(Nsim, clipping)
%
% Simulate the UKF with or without clipping.
pars = batchreactor(Nsim);
Nx = pars.Nx;
Ny = pars.Ny;
Nw = pars.Nw;
Nv = pars.Nv;
pars.Nz = Nx + Nw + Nv; % Augmented state is z = [x; w; v].
Nz = pars.Nz;
% Choose simulation parameters.
Nsim = pars.Nsim;
t = pars.t;
x = pars.xsim;
y = pars.ysim;
Px = pars.P;
Qw = pars.Q;
Rv = pars.R;
% Choose UKF weights.
Nsigma = 1 + 2*Nz;
pars.Nsigma = Nsigma;
pars.lambda = 1; % From Vachhani et. al. (2006). Guarantees Pz > 0.
pars.weights = [2*pars.lambda; ones(2*Nz, 1)]/(2*(Nz + pars.lambda));
W = diag(pars.weights);
% Preallocate variables.
xhat = NaN(Nx, Nsim + 1);
yhat = NaN(Ny, Nsim + 1);
e = NaN(Ny, Nsim + 1);
xhat(:,1) = pars.xhat0; % Poor initial guess
zhat(1:Nx,1) = pars.xhat0;
Pz = blkdiag(Px, Qw, Rv); % Covariance for extended state.
[x_sigma, w_sigma, v_sigma] = get_sigma(Pz, xhat(:,1), pars);
for t = 1:(Nsim + 1)
% Take measurement.
y_sigma = NaN(Ny, Nsigma);
for i = 1:Nsigma
y_sigma(:,i) = full(pars.h(x_sigma(:,i))) + v_sigma(:,i);
end
yhat(:,t) = y_sigma*pars.weights;
e(:,t) = y(:,t) - yhat(:,t);
% Apply correction.
Ex = bsxfun(@minus, x_sigma, xhat(:,t));
Ey = bsxfun(@minus, y_sigma, yhat(:,t));
Exy = Ex*W*Ey';
Eyy = Ey*W*Ey';
L = Exy/Eyy;
Px = Px - L*Exy';
xhat(:,t) = xhat(:,t) + L*e(:,t);
% Stop if at end of simulation.
if t > Nsim
break
end
% Calculate new sigma points, and clip if necessary.
Pz = blkdiag(Px, Qw, Rv);
[x_sigma, w_sigma, v_sigma] = get_sigma(Pz, xhat(:,t), pars);
if clipping
[x_sigma, pars.weights] = ukf_scale(x_sigma, pars);
end
% Simulate sigma points and calculate new xhat and Px.
for i = 1:Nsigma
x_sigma(:,i) = full(pars.f(x_sigma(:,i), w_sigma(:,i)));
end
xhat(:,t + 1) = x_sigma*pars.weights;
Ex = bsxfun(@minus, x_sigma, xhat(:,t + 1));
Px = Ex*W*Ex';
end
% Make a plot.
pars.reactorplot(x, xhat, y, yhat, pars.yclean, pars.Delta);
set(gca(), 'ylim', [-1, 1.5]);
% Form data table.
data = [pars.t', x', xhat', y', yhat'];
|
