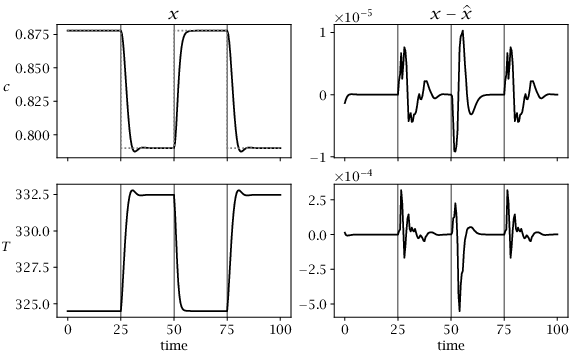
Figure 4.7:
Closed-loop performance of combined nonlinear MHE/MPC with no disturbances. First column shows system states, and second column shows estimation error. Dashed line shows concentration setpoint. Vertical lines indicate times of setpoint changes.
Code for Figure 4.7
Text of the GNU GPL.
main.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27 | % Control of a nonlinear CSTR using nonlinear MPC and MHE.
wmaxes = [0, 0.001, 0.001, 0.01, 0.01, 0.025];
vmaxes = [0, 0.1, 1, 1, 10, 10];
data = cell(size(wmaxes));
% Define helper functions.
% Simulate with varying disturbance sizes.
fig = figure();
ax = zeros(length(wmaxes));
for i = 1:length(ax)
ax(i) = subplot(2, 3, i);
end
for i = 1:length(wmaxes)
data{i} = runcstr(wmaxes(i), vmaxes(i));
axes(ax(i));
plot(data{i}.x(1,:), data{i}.x(2,:), '-ok');
title(sprintf('|w| < %g, |v| < %g', data{i}.wmax, data{i}.vmax));
xlabel('c');
ylabel('T', 'rotation', 0);
axis([0.7, 1, 322, 338]);
end
|
cstrode.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19 | function rhs = cstrode(x, u, pars)
% Nonlinear ODE model for reactor.
c = x(1);
T = x(2);
h = pars.h;
Tc = u(1);
F = pars.F;
F0 = pars.F;
k = pars.k0*exp(-pars.E/T);
rate = k*c;
dcdt = F0*(pars.c0 - c)/(pars.A*h) - rate;
dTdt = F0*(pars.T0 - T)/(pars.A*h) ...
- pars.DeltaH/pars.rhoCp*rate ...
+ 2*pars.U/(pars.r*pars.rhoCp)*(Tc - T);
rhs = [dcdt; dTdt];
|
stagecost.m
| function cost = stagecost(x, u, Deltau, xsp, usp, Q, R, S)
dx = x - xsp;
du = u - usp;
cost = dx'*Q*dx + du'*R*du + Deltau'*S*Deltau;
|
runcstr.m
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229 | function data = runcstr(wmax, vmax)
% data = runcstr(wmax, vmax)
%
% Runs a closed-loop simulation of the CSTR with the given disturbance sizes.
narginchk(2, 2);
mpc = import_mpctools();
% Sizes for the nonlinear system.
Delta = 0.5;
Nx = 2;
Nu = 1;
Ny = 1;
Nw = 1;
Nv = Ny;
small = 1e-5; % Small number.
% Parameters.
pars = struct();
pars.T0 = 350; % K
pars.c0 = 1; % kmol/m^3
pars.r = 0.219; % m
pars.k0 = 7.2e10; % min^-1
pars.E = 8750; % K
pars.U = 54.94; % kJ/(min m^2 K)
pars.rho = 1e3; % kg/m^3
pars.Cp = 0.239; % kJ/(kg K)
pars.DeltaH = -5e4; % kJ/kmol
pars.A = pi()*pars.r.^2;
pars.rhoCp = pars.rho*pars.Cp;
pars.h = 0.659;
pars.F = 0.1;
ode = @(x, u) cstrode(x, u, pars);
cstrsim = mpc.getCasadiIntegrator(ode, Delta, [Nx, Nu], ...
{'x', 'u'}, {'cstr'});
% Steady-state values.
cs = 0.878; % kmol/m^3
Ts = 324.5; % K
Tcs = 300; % K
xs = [cs; Ts];
us = [Tcs];
xlb = [0.1; 250];
xub = [1; 350];
% Simulate a few steps so we actually get dx/dt = 0 at steady state.
for i = 1:10
xs = full(cstrsim(xs, us));
end
cs = xs(1);
Ts = xs(2);
C = [0, 1]; % Can't measure concentration.
ys = C*xs;
CVs = [1]; % Concentration is controlled variable.
G = [1; 0]; % State noise only enters at concentration.
% Bounds.
umax = 0.25*Tcs;
ulb = us - umax;
uub = us + umax;
% Get Kalman Filter weight as a prior.
[A, B] = mpc.getLinearizedModel(ode, {xs, us}, 'Delta', Delta, 'deal', true());
Qw = 0.05;
Rv = 50;
% Make functions for controller, target finder, and estimator.
fprintf('Building controllers.\n');
fmpc = mpc.getCasadiFunc(ode, [Nx, Nu], {'x', 'u'}, ...
'rk4', true(), 'Delta', Delta, 'M', 2, ...
'funcname', 'fnonlin');
fmhe = mpc.getCasadiFunc(@(x, u, w) fmpc(x, u) + G*w, [Nx, Nu, Nw], ...
{'x', 'u', 'w'}, 'funcname', 'fnonlin_w');
fsstarg = mpc.getCasadiFunc(ode, [Nx, Nu], {'x', 'u'}, 'funcname', 'fsstarg');
% Make stage costs.
lcontroller = mpc.getCasadiFunc(@stagecost, ...
{Nx, Nu, Nu, Nx, Nu, [Nx, Nx], [Nu, Nu], [Nu, Nu]}, ...
{'x', 'u', 'Du', 'xsp', 'usp', 'Q', 'R', 'S'}, ...
'funcname', 'l');
Vf = mpc.getCasadiFunc(@(x, xsp, P) (x - xsp)'*P*(x - xsp), ...
{Nx, Nx, [Nx, Nx]}, {'x', 'xsp', 'P'}, 'funcname', 'Vf');
lmhe = mpc.getCasadiFunc(@(w, v, Qinv, Rinv) w'*Qinv*w + v'*Rinv*v, ...
{Nw, Nv, [Nw, Nw], [Nv, Nv]}, ...
{'w', 'v', 'Qinv', 'Rinv'}, 'funcname', 'l');
h = mpc.getCasadiFunc(@(x) C*x, [Nx], {'x'}, 'funcname', 'h');
% Assemble controller, target finder, and estimator.
Ncontroller = 10;
Nmhe = 10;
Q = 0.01*diag(xs.^-2);
R = diag(us.^-2);
[~, P] = dlqr(A, B, Q, R); % For terminal penalty.
S = 10*R; % Rate-of-change penalty.
N = struct('x', Nx, 'u', Nu, 't', Ncontroller);
guess = struct('x', repmat(xs, 1, N.t + 1), 'u', repmat(us, 1, N.t));
lb = struct('x', xlb, 'u', ulb);
ub = struct('x', xub, 'u', uub);
par = struct('xsp', xs, 'usp', us, 'Q', Q, 'R', R, 'P', P, 'S', S, 'uprev', us);
controller = mpc.nmpc('f', fmpc, 'l', lcontroller, 'Vf', Vf, 'N', N, ...
'lb', lb, 'ub', ub, 'guess', guess, 'par', par, ...
'verbosity', 0);
N = struct('x', Nx, 'u', Nu, 'y', Ny);
guess = struct('x', xs, 'u', us, 'y', ys);
lb = struct('x', xlb, 'u', ulb);
ub = struct('x', xub, 'u', uub);
par = struct();
sstarg = mpc.sstarg('f', fsstarg, 'h', h, 'N', N, ...
'lb', lb, 'ub', ub, 'guess', guess, 'par', par, ...
'discretef', false(), 'verbosity', 0);
N = struct('x', Nx, 'u', Nu, 'w', Nw, 'y', Ny, 't', Nmhe);
guess = struct('x', repmat(xs, 1, N.t + 1));
lb = struct('x', xlb);
ub = struct('x', xub);
par = struct('y', repmat(ys, 1, N.t + 1), 'u', repmat(us, 1, N.t), ...
'Qinv', mpc.spdinv(Qw), 'Rinv', mpc.spdinv(Rv));
mhe = mpc.nmhe('f', fmhe, 'h', h, 'l', lmhe, 'N', N, ...
'guess', guess, 'lb', lb, 'ub', ub, 'par', par, ...
'verbosity', 0);
% Closed-loop simulation.
Nsim = 200;
x = NaN(Nx, Nsim + 1);
x(:,1) = xs;
y = NaN(Ny, Nsim + 1);
u = NaN(Nu, Nsim);
rand('state', -1);
urand = @(n, m) 2*rand(n, m) - 1; % Uniform on [-1, 1].
v = vmax*urand(Ny, Nsim + 1);
w = wmax*urand(Nw, Nsim);
mhe.par.y = mhe.par.y + vmax*urand(Ny, Nmhe + 1);
xhat = NaN(Nx, Nsim + 1);
xtarg = NaN(Nx, Nsim);
utarg = NaN(Nu, Nsim);
% Disturbance and setpoint.
xsp = NaN(Nx, Nsim + 1);
t = 0:Nsim;
Nchange = 100;
xsp(CVs,:) = xs(CVs)*(1 - 0.1*(mod(t, Nchange) >= Nchange/2));
xsp(:,end) = xsp(:,end - 1);
% Start loop.
for i = 1:(Nsim + 1)
fprintf('(%3d) ', i);
% Use steady-state target selector.
sstarg.fixvar('x', 1, xsp(CVs,i), CVs);
sstarg.solve();
fprintf('Target: %s, ', sstarg.status);
if ~isequal(sstarg.status, 'Solve_Succeeded')
fprintf('\n');
warning('sstarg failed at time %d!', i);
break
end
xtarg(:,i) = sstarg.var.x;
utarg(:,i) = sstarg.var.u;
% Take measurement and run mhe.
y(:,i) = C*x(:,i) + v(:,i);
mhe.newmeasurement(y(:,i), controller.par.uprev);
mhe.solve();
fprintf('Estimator: %s, ', mhe.status);
if ~isequal(mhe.status, 'Solve_Succeeded')
fprintf('\n');
warning('mhe failed at time %d!', i);
break
end
xhat(:,i) = mhe.var.x(:,end);
mhe.saveguess();
% Stop if at last time.
if i == Nsim + 1
fprintf('Done\n');
break
end
% Apply control law.
controller.fixvar('x', 1, xhat(:,i));
controller.par.xsp = xtarg(:,i);
controller.par.usp = utarg(:,i);
controller.solve();
fprintf('Controller: %s, ', controller.status);
if ~isequal(controller.status, 'Solve_Succeeded')
fprintf('\n');
warning('controller failed at time %d', i);
break
end
u(:,i) = controller.var.u(:,1);
controller.saveguess();
controller.par.uprev = u(:,i); % Save previous u.
% Evolve plant.
x(:,i + 1) = full(cstrsim(x(:,i), u(:,i))) + G*w(:,i);
fprintf('\n');
end
% Make a plot.
t = Delta*(0:Nsim);
plottitle = sprintf('|w| < %g, |v| < %g', wmax, vmax);
style = struct('fig', figure(), 'marker', '', 't', t);
mpc.mpcplot('x', x, 'u', u, 'xnames', {'c', 'T'}, ...
'unames', {'T_c'}, 'title', plottitle, 'legend', 'Actual', ...
'**', style);
mpc.mpcplot('x', xhat, 'u', NaN(Nu, Nsim), 'color', 'b', 'linestyle', '--', ...
'legend', 'Estimated', '**', style);
mpc.mpcplot('x', xsp, 'u', NaN(Nu, Nsim), 'color', 'r', 'linestyle', ':', ...
'legend', 'Setpoint', '**', style);
mpc.mpcplot('x', [NaN(1, Nsim + 1); y], 'u', NaN(size(u)), 'color', 'g', ...
'legend', 'Measurement', '**', style);
% Store data.
data = struct('x', x, 'u', u, 'y', y, 'xhat', xhat, 'xsp', xsp, 't', t, ...
'v', v, 'w', w, 'vmax', vmax, 'wmax', wmax);
|
