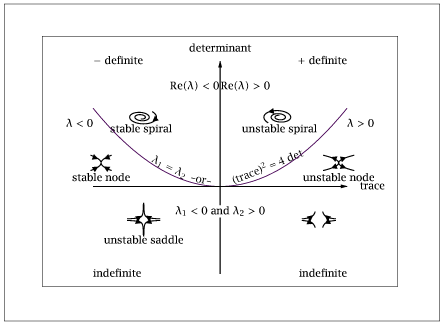
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106 | import numpy as np
import matplotlib.pyplot as plt
from scipy.linalg import expm
# Axes and (trace)^2 = 4 det line
x = np.linspace(-5, 5, 101)
y = x**2 / 4
quad = np.column_stack((x, y))
lines = np.array([
[-5, 0, 0, -7, 0, -1.5],
[5, 0, 0, -2.5, 0, 10]
])
# Stable spiral
T = -0.2
D = (T**2 + 4) / 4
A = np.array([[0, np.sqrt(D)], [-np.sqrt(D), T]])
nts = 101
time = np.linspace(0, 15, nts)
x0 = np.array([1, 1])
xssp = np.zeros((nts, 2))
for i in range(len(time)):
xssp[i] = expm(A * time[i]) @ x0
# Unstable spiral (reverse time)
x0 = np.array([-1, -1])
xusp = np.zeros((nts, 2))
for i in range(len(time)):
xusp[i] = expm(A * time[i]) @ x0
xusp = xusp[::-1]
# Stable node
A = np.array([[-0.5, 0], [0, -0.25]])
x0 = np.array([1, 1])
x = np.zeros((nts, 2))
for i in range(len(time)):
x[i] = expm(A * time[i]) @ x0
x1, x2 = x[:, 0], x[:, 1]
xsno = np.column_stack((x, -x1, x2, -x1, -x2, x1, -x2))
# Unstable saddle, trace < 0
A = np.array([[-0.3, 0], [0, 0.2]])
x0 = np.array([-1, 0.1])
x = np.zeros((nts, 2))
for i in range(len(time)):
x[i] = expm(A * time[i]) @ x0
x1, x2 = x[:, 0], x[:, 1]
xusa = np.column_stack((x, -x1, x2, -x1, -x2, x1, -x2))
# Another unstable saddle, trace > 0
A = np.array([[-0.1, 0], [0, 0.15]])
x0 = np.array([-1, 0.1])
x = np.zeros((nts, 2))
for i in range(len(time)):
x[i] = expm(A * time[i]) @ x0
x1, x2 = x[:, 0], x[:, 1]
xusat = np.column_stack((x, -x1, x2, -x1, -x2, x1, -x2))
# Plotting
plt.figure(figsize=(12, 10))
plt.plot(quad[:, 0], quad[:, 1], 'k-')
plt.plot(lines[:, 0], lines[:, 1], 'k-')
plt.plot(lines[:, 2], lines[:, 3], 'k-')
plt.plot(lines[:, 4], lines[:, 5], 'k-')
plt.plot(xssp[:, 0], xssp[:, 1], 'r-', label='Stable Spiral')
plt.plot(xusp[:, 0], xusp[:, 1], 'b-', label='Unstable Spiral')
for i in range(0, xsno.shape[1], 2):
plt.plot(xsno[:, i], xsno[:, i+1], 'g-')
plt.plot([], [], 'g-', label='Stable Node')
for i in range(0, xusa.shape[1], 2):
plt.plot(xusa[:, i], xusa[:, i+1], 'm-')
plt.plot([], [], 'm-', label='Unstable Saddle (trace < 0)')
for i in range(0, xusat.shape[1], 2):
plt.plot(xusat[:, i], xusat[:, i+1], 'c-')
plt.plot([], [], 'c-', label='Unstable Saddle (trace > 0)')
plt.title('Complicated Planar Dynamics')
plt.xlabel('x')
plt.ylabel('y')
plt.legend()
plt.grid(True)
plt.axis('equal')
plt.xlim(-5, 5)
plt.ylim(-5, 5)
plt.show(block=False)
# Save data to .dat file
with open('jimphase.dat', 'w') as f:
np.savetxt(f, quad, fmt='%f', header="quad")
f.write("\n\n")
np.savetxt(f, lines, fmt='%f', header="lines")
f.write("\n\n")
np.savetxt(f, xssp, fmt='%f', header="xssp")
f.write("\n\n")
np.savetxt(f, xusp, fmt='%f', header="xusp")
f.write("\n\n")
np.savetxt(f, xsno, fmt='%f', header="xsno")
f.write("\n\n")
np.savetxt(f, xusa, fmt='%f', header="xusa")
f.write("\n\n")
np.savetxt(f, xusat, fmt='%f', header="xusat")
|