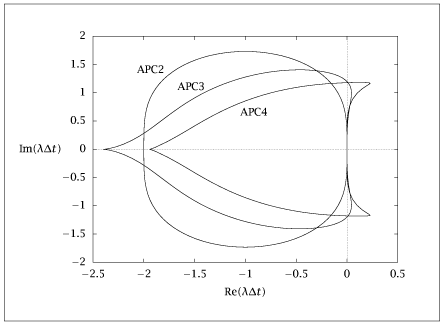
import numpy as np
from scipy.optimize import fsolve
import matplotlib.pyplot as plt
# Setting grid points
axisbox = [-1.5, 1.5, -1, 1]
xa, xb, ya, yb = axisbox
nhalf = 50
npts = 2*nhalf+1
theta = np.linspace(0, 4*np.pi, npts)
lamvec = np.exp(1j*theta)
# predictor coefficients, 1st, 2nd, 3rd, 4th order
p = [[1, 0, 0, 0],
[1.5, -0.5, 0, 0],
[23/12, -4/3, 5/12, 0],
[55/24, -59/24, 37/24, -9/24]]
# corrector coefficients, 1st, 2nd, 3rd, 4th order
c = [[1, 0, 0, 0],
[0.5, 0.5, 0, 0],
[5/12, 8/12, -1/12, 0],
[9/24, 19/24, -5/24, 1/24]]
def characteristic(w, PCs, lam):
cc = c[PCs-1]
pp = p[PCs-2] # note, the predictor order is one less than corrector
a = np.zeros(4, dtype=complex)
a[0] = 1 + w * (cc[0] * (1 + w * pp[0]) + cc[1])
a[1] = w * (cc[0] * w * pp[1] + cc[2])
a[2] = w * (cc[0] * w * pp[2] + cc[3])
a[3] = w * cc[0] * w * pp[3]
order = len(a)
G = np.zeros([order, order], dtype=complex)
G[0,:] = a
G[1:,:-1] = np.eye(order-1)
G -= lam * np.eye(order)
return np.linalg.det(G)
def func(w, PCs, lam):
return [characteristic(w[0] + 1j*w[1], PCs, lam).real,
characteristic(w[0] + 1j*w[1], PCs, lam).imag]
PCsvec = [2, 3, 4]
npcs = len(PCsvec)
wsol = [np.zeros(npts, dtype=complex) for _ in range(npcs)]
for j, PCs in enumerate(PCsvec):
w0 = 0 + 0j
for k, lam in enumerate(lamvec):
sol = fsolve(func, [w0.real, w0.imag], args=(PCs, lam), full_output=True)
if sol[2] == 1:
w = sol[0][0] + 1j*sol[0][1]
wsol[j][k] = w
w0 = 0.95*w
else:
print(f"Warning: fsolve did not converge for j={j}, k={k}")
print(sol)
wsav = wsol.copy()
# Check for intersecting loops
for ind in range(npcs):
for k in range(len(wsol[ind]) - 1):
z = np.array([wsol[ind].real, wsol[ind].imag])
iloop = 0
for j in range(k+2, len(wsol[ind])-1):
A = np.column_stack([z[:,k]-z[:,k+1], z[:,j+1]-z[:,j]])
if np.linalg.det(A) != 0:
lam = np.linalg.solve(A, z[:,j+1]-z[:,k+1])
if np.all((0 <= lam) & (lam <= 1)):
iloop = list(range(k+1, j+1))
zint = lam[0]*z[:,k] + (1-lam[0])*z[:,k+1]
break
if iloop:
zcp = complex(zint[0], zint[1])
wsol[ind][iloop[0]] = zcp
wsol[ind] = np.delete(wsol[ind], iloop[1:])
plt.figure()
for j in range(npcs):
plt.plot(wsol[j].real, wsol[j].imag, '-o')
plt.show(block=False)
# Prepare data for saving
# Save to APC_stability.dat file
with open("APC_stability.dat", "w") as f:
for j in range(npcs):
store = np.column_stack((wsol[j].real, wsol[j].imag))
np.savetxt(f, store, fmt='%.6f', header=f"predictor/corrector order:{j+1}/{j+2}")
f.write("\n\n")