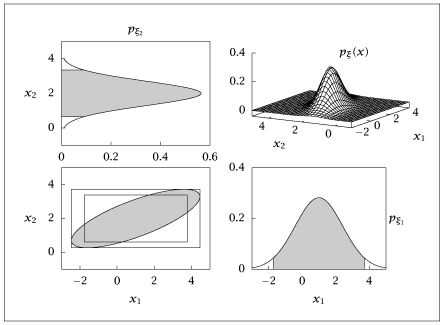
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142 | import numpy as np
from numpy.linalg import inv
from scipy.stats import chi2
from scipy.linalg import sqrtm
import matplotlib.pyplot as plt
from misc import ellipse
# Set parameters
m = np.array([[1], [2]])
n = len(m)
P = np.array([[2, 0.75], [0.75, 0.5]])
nsam = 1000
np.random.seed(0)
# Generate samples
x = np.tile(m, (1, nsam)) + sqrtm(P) @ np.random.randn(n, nsam)
# Compute the 95% confidence interval ellipse
alpha = 0.95
A = inv(P)
b = chi2.ppf(alpha, n)
xe, ye, *_ = ellipse(A, b, 100, m)
# Count how many samples are outside the ellipse
e = x - np.tile(m, (1, nsam))
outside_ellipse = np.sum(np.diag((e.T @ A) @ e) > b) / nsam
# Compute the 95% bounding box
xd = np.sqrt(b * np.diag(P))
bbox = np.array([
[-xd[0], -xd[1]],
[xd[0], -xd[1]],
[xd[0], xd[1]],
[-xd[0], xd[1]],
[-xd[0], -xd[1]]
])
bbox = bbox + np.tile(m.T, (5, 1))
# Count samples outside the bounding box
z = x - np.tile(m, (1, nsam))
outside_bbox = sum( sum(np.abs(z) > xd[:,None]) > 0 ) / nsam
# Compute 95% marginal intervals
b2 = chi2.ppf(alpha, 1)
xd2 = np.sqrt(b2 * np.diag(P))
mbox = np.array([
[-xd2[0], -xd2[1]],
[xd2[0], -xd2[1]],
[xd2[0], xd2[1]],
[-xd2[0], xd2[1]],
[-xd2[0], -xd2[1]]
])
mbox = mbox + np.tile(m.T, (5, 1))
# Count samples outside the marginal box
outside_mbox = sum( sum(np.abs(z) > xd2[:,None]) > 0 ) / nsam
# Prepare mtab data (xplot, fx for both components)
xrange = np.round([np.min(x, axis=1), np.max(x, axis=1)]).astype(int).T
nplot = 100
xplot = np.linspace(xrange[:,0], xrange[:,1], nplot).T
z = xplot - m
diagP = np.diag(P)[:,None];
fx = np.exp( -(1/2) * (z / diagP * z)) / np.sqrt(2*np.pi*diagP)
# compute closed curves for using filledcurve in gnuplot
ind = abs(z) <= xd2[:,None]
xp1 = xplot[0,:]
xp2 = xplot[1,:]
fx1 = fx[0,:]
fx2 = fx[1,:]
ind1 = ind[0,:]
ind2 = ind[1,:]
fill1 = np.row_stack( (xp1[ind1], fx1[ind1]) )
fill1 = np.column_stack( (np.row_stack( (fill1[0,0], 0) ), fill1, np.row_stack( (fill1[0,-1], 0) ) ) ).T
fill2 = np.row_stack( (xp2[ind2], fx2[ind2]) )
fill2 = np.column_stack( (np.row_stack( (fill2[0,0], 0) ), fill2, np.row_stack( (fill2[0,-1], 0) ) ) ).T
nbins = 20
# Histogram for x[0,:]
n1, bins1 = np.histogram(x[0,:], bins=nbins, density=True)
bin_centers1 = 0.5 * (bins1[:-1] + bins1[1:])
# make bar plot data for gnuplot
barx1 = np.kron(bins1,[1,1,1])[1:-1]
bary1 = np.append(np.kron(n1, [0, 1, 1]), 0)
# Histogram for x[1,:]
n2, bins2 = np.histogram(x[1,:], bins=nbins, density=True)
bin_centers2 = 0.5 * (bins2[:-1] + bins2[1:])
# make bar plot data for gnuplot
barx2 = np.kron(bins2,[1,1,1])[1:-1]
bary2 = np.append(np.kron(n2, [0, 1, 1]), 0)
plt.figure()
plt.plot(x[0,:], x[1,:], 'o', mfc='none')
plt.plot(xe, ye)
plt.plot(bbox[:,0], bbox[:,1])
plt.plot(mbox[:,0], mbox[:,1])
plt.figure()
plt.plot(barx1, bary1, label='Histogram x[1,:]')
plt.figure()
plt.plot(barx2, bary2, label='Histogram x[2,:]')
plt.figure()
plt.plot (xplot[0,:], fx[0,:])
plt.plot (xplot[1,:], fx[1,:])
plt.show(block=False)
samtab = x.T
eltab = np.column_stack([xe, ye])
mtab = np.column_stack( (xplot.T, fx.T) )
# Create the .dat file with correct formatting for Gnuplot
with open("confmarg.dat", "w") as f:
# Write sample matrix (samtab)
np.savetxt(f, samtab, fmt='%f', header="samtab")
f.write("\n\n")
# Write ellipse data (eltab)
np.savetxt(f, eltab, fmt='%f', header="eltab")
f.write("\n\n")
# Write ellipse data (eltab)
np.savetxt(f, mtab, fmt='%f', header="mtab")
f.write("\n\n")
# Write bounding box data (bbox)
np.savetxt(f, bbox, fmt='%f', header="bbox")
f.write("\n\n")
# Write marginal box data (mbox)
np.savetxt(f, mbox, fmt='%f', header="mbox")
f.write("\n\n")
# Write fill1 and fill2 data
np.savetxt(f, fill1, fmt='%f', header="fill1")
f.write("\n\n")
np.savetxt(f, fill2, fmt='%f', header="fill2")
|