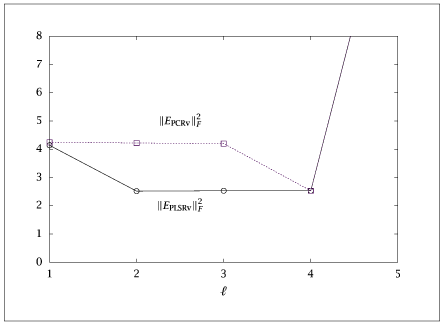
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121 | import numpy as np
import matplotlib.pyplot as plt
from scipy.linalg import svd
n = 100
p = 2
q = 5
#np.random.seed(1)
np.random.seed(0)
# True parameters
B = np.vstack((np.zeros((3, 2)), [-1, 1], [0, 0]))
# Generate data
X = np.random.rand(n, q-1)
xq = X @ np.ones((q-1, 1)) / np.sqrt(q-1) + 0.01 * np.random.rand(n, 1)
X = np.hstack((X, xq))
Y = X @ B + 0.1 * np.random.rand(n, p)
# Cross-validation data
X2 = np.random.rand(n, q)
Y2 = X2 @ B + 0.1 * np.random.rand(n, p)
Xdat = np.vstack((X, X2))
Ydat = np.vstack((Y, Y2))
# Save the data in low precision
with open('pca_pls_data.dat', 'w') as myfile:
myfile.write('# x1 x2 x3 x4 x5 y1 y2\n')
for i in range(2 * n):
line = ' '.join(f'{val:8.3f}' for val in np.hstack((Xdat[i], Ydat[i])))
myfile.write(f'{line}\n')
# Reload data to ensure consistency
Data = np.loadtxt('pca_pls_data.dat', skiprows=1)
X, Y = Data[:n, :q], Data[:n, q:q+p]
X2, Y2 = Data[n:, :q], Data[n:, q:q+p]
# Full least squares
Bls = np.linalg.inv(X.T @ X) @ X.T @ Y
# PCR or SVD
U, S, Vt = svd(X, full_matrices=False)
V = Vt.T
Bsvd = [np.zeros((q, p)) for _ in range(q)]
Ysvd = [np.zeros((n, p)) for _ in range(q)]
errsvd = np.zeros(q)
for l in range(q):
Ul, Sl, Vl = U[:, :l+1], np.diag(S[:l+1]), V[:, :l+1]
Bsvd[l] = Vl @ np.linalg.inv(Sl) @ Ul.T @ Y
Ysvd[l] = X @ Bsvd[l]
errsvd[l] = np.linalg.norm(Y - Ysvd[l])
# PLS algorithm
U, T, P, Q = [], [], [], []
E, F = X.copy(), Y.copy()
Bpls = [np.zeros((q, p)) for _ in range(q)]
Ypls = [np.zeros((n, p)) for _ in range(q)]
errpls = np.zeros(q)
for i in range(q):
UU, SS, VVt = svd(E.T @ F, full_matrices=False)
u, v = UU[:, 0:1], VVt.T[:, 0:1]
t = E @ u
t = t / np.linalg.norm(t)
pp = E.T @ t
qq = F.T @ t
E = E - t @ pp.T
F = F - t @ qq.T
U.append(u)
T.append(t)
P.append(pp)
Q.append(qq)
R = np.hstack(U) @ np.linalg.inv(np.hstack(P).T @ np.hstack(U))
Bpls[i] = R @ np.hstack(Q).T
Ypls[i] = X @ Bpls[i]
errpls[i] = np.linalg.norm(Y - Ypls[i])
plt.figure()
plt.plot(range(q), errsvd, '-x')
plt.plot(range(q), errpls, '-o')
# Save results in correct format for gnuplot
tab1 = np.column_stack((np.arange(1, q+1), errsvd))
tab2 = np.column_stack((np.arange(1, q+1), errpls))
tab3 = np.hstack((Y, np.hstack(Ysvd)))
tab4 = np.hstack((Y, np.hstack(Ypls)))
tab5 = np.column_stack((np.arange(1, q+1), [np.linalg.norm(Y2 - X2 @ Bsvd[i]) for i in range(q)]))
tab6 = np.column_stack((np.arange(1, q+1), [np.linalg.norm(Y2 - X2 @ Bpls[i]) for i in range(q)]))
tab7 = np.hstack((Y2, np.hstack([X2 @ Bsvd[i] for i in range(q)])))
tab8 = np.hstack((Y2, np.hstack([X2 @ Bpls[i] for i in range(q)])))
# Concatenate tables into a single file
with open('pca_pls.dat', 'w') as f:
f.write("# tab1 (errors for SVD)\n")
np.savetxt(f, tab1, fmt='%f')
f.write("\n\n")
f.write("# tab2 (errors for PLS)\n")
np.savetxt(f, tab2, fmt='%f')
f.write("\n\n")
f.write("# tab3 (Y and Ysvd)\n")
np.savetxt(f, tab3, fmt='%f')
f.write("\n\n")
f.write("# tab4 (Y and Ypls)\n")
np.savetxt(f, tab4, fmt='%f')
f.write("\n\n")
f.write("# tab5 (cross-validation errors for SVD)\n")
np.savetxt(f, tab5, fmt='%f')
f.write("\n\n")
f.write("# tab6 (cross-validation errors for PLS)\n")
np.savetxt(f, tab6, fmt='%f')
f.write("\n\n")
f.write("# tab7 (cross-validation Y2 and Y2svd)\n")
np.savetxt(f, tab7, fmt='%f')
f.write("\n\n")
f.write("# tab8 (cross-validation Y2 and Y2pls)\n")
np.savetxt(f, tab8, fmt='%f')
|