import numpy as np
import matplotlib.pyplot as plt
from scipy.integrate import solve_ivp
from scipy.interpolate import interp1d
# Parameters
nmolec = 400
k1 = 1
k2 = 1
k = [k1 / nmolec, k2]
stoi = np.array([[-1, -1, 1], [1, 1, -1]]).T # stoichiometry matrix
nrxs, nspec = stoi.shape
# Initial conditions for three species A, B, C
x = np.zeros((3, nmolec * 4 + 1))
x[:, 0] = [nmolec, 0.9 * nmolec, 0] # A, B, C initial concentrations
time = np.zeros(nmolec * 4 + 1)
# Seed for reproducibility
np.random.seed(0)
# Gillespie algorithm simulation
for n in range(nmolec * 4):
r = [k[0] * x[0, n] * x[1, n], k[1] * x[2, n]]
rtot = sum(r)
p = np.random.rand(2)
tau = -np.log(p[0]) / rtot
time[n + 1] = time[n] + tau
# Determine which reaction occurs
rcum = 0
m = -1
while rcum <= p[1] * rtot:
m += 1
rcum += r[m]
x[:, n + 1] = x[:, n] + stoi[:, m]
# Scale the results
xscale = x / x[0, 0]
# Deterministic comparison using ODE
ca0, cb0, cc0 = 1.0, 0.9, 0.0
timedet = np.concatenate(([0], np.logspace(-4, 1, 100)))
p = {'ca0': ca0, 'cb0': cb0, 'cc0': cc0, 'k1det': k1, 'k2det': k2}
# Define the ODE function
def f(t, x):
ca = x[0]
cb = ca - p['ca0'] + p['cb0']
cc = p['cc0'] + p['ca0'] - ca
dca_dt = -p['k1det'] * ca * cb + p['k2det'] * cc
return [dca_dt]
# Solve the ODE
ode_sol = solve_ivp(f, [timedet[0], timedet[-1]], [ca0], t_eval=timedet, method='LSODA')
ca = ode_sol.y[0]
cb = ca - ca0 + cb0
cc = cc0 + ca0 - ca
# Interpolate deterministic results to match stochastic time points
ca_interp = interp1d(timedet, ca, kind='linear', fill_value="extrapolate")(time)
cb_interp = interp1d(timedet, cb, kind='linear', fill_value="extrapolate")(time)
cc_interp = interp1d(timedet, cc, kind='linear', fill_value="extrapolate")(time)
# Prepare the table for saving
table = np.column_stack((time, xscale.T, ca_interp, cb_interp, cc_interp))
# Save the data to a file
with open("stochnon.dat", "w") as f:
np.savetxt(f, table, fmt='%f', header="time A_sto B_sto C_sto A_det B_det C_det")
# Plot the results
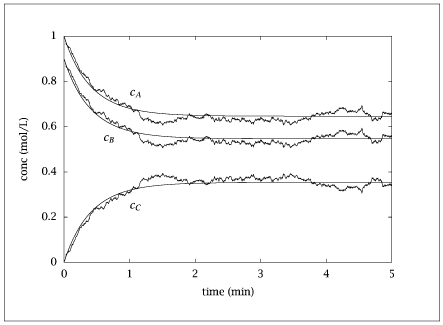
plt.figure()
plt.plot(table[:, 0], table[:, 1:4], label=['Stochastic A', 'Stochastic B', 'Stochastic C'])
plt.plot(time, ca_interp, label='Deterministic A', linestyle='--')
plt.plot(time, cb_interp, label='Deterministic B', linestyle='--')
plt.plot(time, cc_interp, label='Deterministic C', linestyle='--')
plt.xlabel('Time')
plt.ylabel('Concentration (scaled)')
plt.legend()
plt.title('Stochastic vs Deterministic Simulation')
plt.axis([0, 5, 0, 1])
plt.grid()
plt.show(block=False)